Note
Go to the end to download the full example code
PyVista 网格集成#
在 MAPDL 中对 pyvista 生成的网格进行模态分析。
import os
import tempfile
from ansys.mapdl.reader import save_as_archive
import pyvista as pv
from ansys.mapdl.core import launch_mapdl
# launch MAPDL as a service
mapdl = launch_mapdl(loglevel="ERROR")
# 在 XY 平面上创建一个以 (0, 0, 0) 为中心的简单平面网格
mesh = pv.Plane(i_resolution=100, j_resolution=100)
mesh.plot(color="w", show_edges=True)

将网格写入临时文件
archive_filename = os.path.join(tempfile.gettempdir(), "tmp.cdb")
save_as_archive(archive_filename, mesh)
# 读取存档文件
response = mapdl.cdread("db", archive_filename)
mapdl.prep7()
print(mapdl.shpp("SUMM"))
SUMMARIZE SHAPE TESTING FOR ALL SELECTED ELEMENTS
------------------------------------------------------------------------------
<<<<<< SHAPE TESTING SUMMARY >>>>>>
<<<<<< FOR ALL SELECTED ELEMENTS >>>>>>
------------------------------------------------------------------------------
--------------------------------------
| Element count 10000 SHELL181 |
--------------------------------------
Test Number tested Warning count Error count Warn+Err %
---- ------------- ------------- ----------- ----------
Aspect Ratio 10000 0 0 0.00 %
Parallel Deviation 10000 0 0 0.00 %
Maximum Angle 10000 0 0 0.00 %
Jacobian Ratio 10000 0 0 0.00 %
Warping Factor 10000 0 0 0.00 %
Any 10000 0 0 0.00 %
------------------------------------------------------------------------------
指定壳单元厚度
mapdl.sectype(1, "shell")
mapdl.secdata(0.01)
mapdl.emodif("ALL", "SECNUM", 1)
MODIFY ALL SELECTED ELEMENTS TO HAVE SECN = 1
规定材料属性 使用 AISI 5000 系列钢材的近似值
mapdl.units("SI") # not necessary, but helpful for book keeping
mapdl.mp("EX", 1, 200e9) # Elastic moduli in Pa (kg/(m*s**2))
mapdl.mp("DENS", 1, 7800) # Density in kg/m3
mapdl.mp("NUXY", 1, 0.3) # Poissons Ratio
mapdl.emodif("ALL", "MAT", 1)
MODIFY ALL SELECTED ELEMENTS TO HAVE MAT = 1
运行无约束模态分析 for the first 20 modes above 1 Hz
mapdl.modal_analysis(nmode=20, freqb=1)
# 您还可以这样运行:
# mapdl.run('/SOLU')
# mapdl.antype('MODAL') # default NEW
# mapdl.modopt('LANB', 20, 1)
# mapdl.solve()
*** NOTE *** CP = 7.734 TIME= 23:59:26
The automatic domain decomposition logic has selected the MESH domain
decomposition method with 2 processes per solution.
***** MAPDL SOLVE COMMAND *****
*** NOTE *** CP = 7.734 TIME= 23:59:26
There is no title defined for this analysis.
*** NOTE *** CP = 7.734 TIME= 23:59:26
No modes are being expanded (MXPAND command) and therefore the element
results will not be written to the mode file. If you do not intend to
use mode selection and for more efficient calculation of element
results in the expansion pass of any downstream mode superposition
analyses, expand all modes during the modal analysis.
*** SELECTION OF ELEMENT TECHNOLOGIES FOR APPLICABLE ELEMENTS ***
---GIVE SUGGESTIONS ONLY---
ELEMENT TYPE 4 IS SHELL181. IT IS ASSOCIATED WITH ELASTOPLASTIC
MATERIALS ONLY. KEYOPT(8)=2 IS SUGGESTED AND KEYOPT(3)=2 IS SUGGESTED FOR
HIGHER ACCURACY OF MEMBRANE STRESSES; OTHERWISE, KEYOPT(3)=0 IS SUGGESTED.
*** MAPDL - ENGINEERING ANALYSIS SYSTEM RELEASE 2023 R1 23.1 ***
Ansys Mechanical Enterprise
20120530 VERSION=WINDOWS x64 23:59:26 JAN 25, 2024 CP= 7.734
S O L U T I O N O P T I O N S
PROBLEM DIMENSIONALITY. . . . . . . . . . . . .3-D
DEGREES OF FREEDOM. . . . . . UX UY UZ ROTX ROTY ROTZ
ANALYSIS TYPE . . . . . . . . . . . . . . . . .MODAL
EXTRACTION METHOD. . . . . . . . . . . . . .BLOCK LANCZOS
NUMBER OF MODES TO EXTRACT. . . . . . . . . . . 20
SHIFT POINT . . . . . . . . . . . . . . . . . . 1.0000
GLOBALLY ASSEMBLED MATRIX . . . . . . . . . . .SYMMETRIC
*** WARNING *** CP = 7.766 TIME= 23:59:26
No constraints have been defined using the D command.
*** NOTE *** CP = 7.766 TIME= 23:59:26
The step data was checked and warning messages were found.
Please review output or errors file (
C:\Users\ff\AppData\Local\Temp\ansys_orzczjzgnq\file0.err ) for these
warning messages.
*** NOTE *** CP = 7.766 TIME= 23:59:26
The conditions for direct assembly have been met. No .emat or .erot
files will be produced.
D I S T R I B U T E D D O M A I N D E C O M P O S E R
...Number of elements: 10000
...Number of nodes: 10201
...Decompose to 2 CPU domains
...Element load balance ratio = 1.000
L O A D S T E P O P T I O N S
LOAD STEP NUMBER. . . . . . . . . . . . . . . . 1
THERMAL STRAINS INCLUDED IN THE LOAD VECTOR . . YES
*********** PRECISE MASS SUMMARY ***********
TOTAL RIGID BODY MASS MATRIX ABOUT ORIGIN
Translational mass | Coupled translational/rotational mass
78.000 0.0000 0.0000 | 0.0000 0.0000 0.11102E-15
0.0000 78.000 0.0000 | 0.0000 0.0000 0.35527E-14
0.0000 0.0000 78.000 | -0.66613E-15 -0.35527E-14 0.0000
------------------------------------------ | ------------------------------------------
| Rotational mass (inertia)
| 6.5006 0.34694E-15 0.0000
| 0.17000E-15 6.5006 0.0000
| 0.0000 0.0000 12.999
TOTAL MASS = 78.000
The mass principal axes coincide with the global Cartesian axes
CENTER OF MASS (X,Y,Z)= 0.45548E-16 -0.14234E-17 0.0000
TOTAL INERTIA ABOUT CENTER OF MASS
6.5006 0.34694E-15 0.0000
0.34694E-15 6.5006 0.0000
0.0000 0.0000 12.999
The inertia principal axes coincide with the global Cartesian axes
*** MASS SUMMARY BY ELEMENT TYPE ***
TYPE MASS
4 78.0000
Range of element maximum matrix coefficients in global coordinates
Maximum = 749504998 at element 9585.
Minimum = 749503915 at element 9596.
*** ELEMENT MATRIX FORMULATION TIMES
TYPE NUMBER ENAME TOTAL CP AVE CP
4 10000 SHELL181 0.281 0.000028
Time at end of element matrix formulation CP = 7.96875.
BLOCK LANCZOS CALCULATION OF UP TO 20 EIGENVECTORS.
NUMBER OF EQUATIONS = 61206
MAXIMUM WAVEFRONT = 26
MAXIMUM MODES STORED = 20
MINIMUM EIGENVALUE = 0.10000E+01
MAXIMUM EIGENVALUE = 0.10000E+31
Process memory allocated for solver = 66.697 MB
Process memory required for in-core solution = 63.679 MB
Process memory required for out-of-core solution = 30.081 MB
Total memory allocated for solver = 132.847 MB
Total memory required for in-core solution = 126.837 MB
Total memory required for out-of-core solution = 58.694 MB
*** NOTE *** CP = 8.328 TIME= 23:59:27
The Distributed Sparse Matrix Solver used by the Block Lanczos
eigensolver is currently running in the in-core memory mode. This
memory mode uses the most amount of memory in order to avoid using the
hard drive as much as possible, which most often results in the
fastest solution time. This mode is recommended if enough physical
memory is present to accommodate all of the solver data.
Total memory available for LANCZOS = 39.727852 GB
Process memory required for in-core LANCZOS Workspace = 122.022560 MB
Process memory required for out-of-core LANCZOS Workspace = 3.376808 MB
Total memory required for in-core LANCZOS Workspace = 241.530350 MB
Total memory required for out-of-core LANCZOS Workspace = 6.693741 MB
Lanczos Memory Mode : INCORE
*** MAPDL - ENGINEERING ANALYSIS SYSTEM RELEASE 2023 R1 23.1 ***
Ansys Mechanical Enterprise
20120530 VERSION=WINDOWS x64 23:59:28 JAN 25, 2024 CP= 8.609
*** FREQUENCIES FROM BLOCK LANCZOS ITERATION ***
MODE FREQUENCY (HERTZ)
FREQUENCY RANGE REQUESTED= 20 MODES ABOVE 1.00000 HERTZ
1 32.74664192913
2 47.77804288706
3 59.17108298309
4 84.60790505091
5 84.60790505194
6 148.8927748644
7 148.8927748649
8 154.6519861755
9 168.3840411456
10 187.7790667336
11 255.9751634910
12 255.9751634923
13 285.2444050784
14 298.2943388358
15 319.8023401629
16 319.8023401639
17 370.6023621795
18 391.8457651354
19 409.1039880837
20 482.6516135507
*** MAPDL - ENGINEERING ANALYSIS SYSTEM RELEASE 2023 R1 23.1 ***
Ansys Mechanical Enterprise
20120530 VERSION=WINDOWS x64 23:59:28 JAN 25, 2024 CP= 8.609
***** PARTICIPATION FACTOR CALCULATION ***** X DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
***** PARTICIPATION FACTOR CALCULATION ***** Y DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
***** PARTICIPATION FACTOR CALCULATION ***** Z DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
***** PARTICIPATION FACTOR CALCULATION *****ROTX DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
***** PARTICIPATION FACTOR CALCULATION *****ROTY DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
***** PARTICIPATION FACTOR CALCULATION *****ROTZ DIRECTION
CUMULATIVE RATIO EFF.MASS
MODE FREQUENCY PERIOD PARTIC.FACTOR RATIO EFFECTIVE MASS MASS FRACTION TO TOTAL MASS
1 32.7466 0.30537E-01 0.0000 0.000000 0.00000 0.00000 0.00000
2 47.7780 0.20930E-01 0.0000 0.000000 0.00000 0.00000 0.00000
3 59.1711 0.16900E-01 0.0000 0.000000 0.00000 0.00000 0.00000
4 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
5 84.6079 0.11819E-01 0.0000 0.000000 0.00000 0.00000 0.00000
6 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
7 148.893 0.67162E-02 0.0000 0.000000 0.00000 0.00000 0.00000
8 154.652 0.64661E-02 0.0000 0.000000 0.00000 0.00000 0.00000
9 168.384 0.59388E-02 0.0000 0.000000 0.00000 0.00000 0.00000
10 187.779 0.53254E-02 0.0000 0.000000 0.00000 0.00000 0.00000
11 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
12 255.975 0.39066E-02 0.0000 0.000000 0.00000 0.00000 0.00000
13 285.244 0.35058E-02 0.0000 0.000000 0.00000 0.00000 0.00000
14 298.294 0.33524E-02 0.0000 0.000000 0.00000 0.00000 0.00000
15 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
16 319.802 0.31269E-02 0.0000 0.000000 0.00000 0.00000 0.00000
17 370.602 0.26983E-02 0.0000 0.000000 0.00000 0.00000 0.00000
18 391.846 0.25520E-02 0.0000 0.000000 0.00000 0.00000 0.00000
19 409.104 0.24444E-02 0.0000 0.000000 0.00000 0.00000 0.00000
20 482.652 0.20719E-02 0.0000 0.000000 0.00000 0.00000 0.00000
*** MAPDL BINARY FILE STATISTICS
BUFFER SIZE USED= 16384
8.625 MB WRITTEN ON ASSEMBLED MATRIX FILE: file0.full
1.125 MB WRITTEN ON MODAL MATRIX FILE: file0.mode
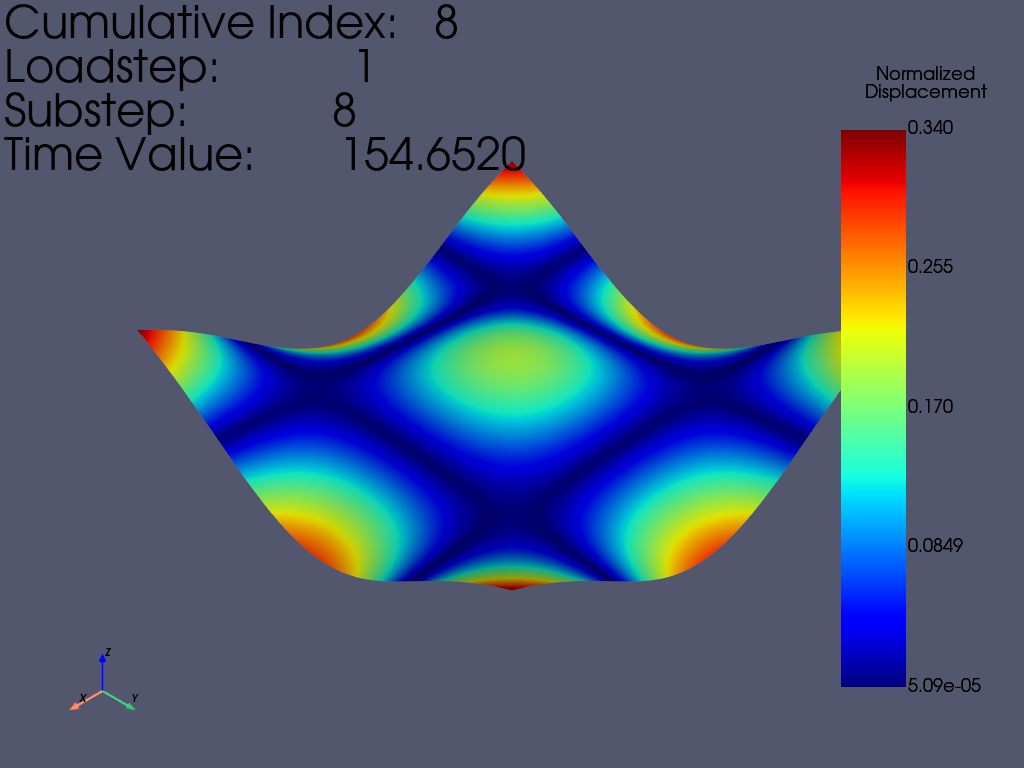
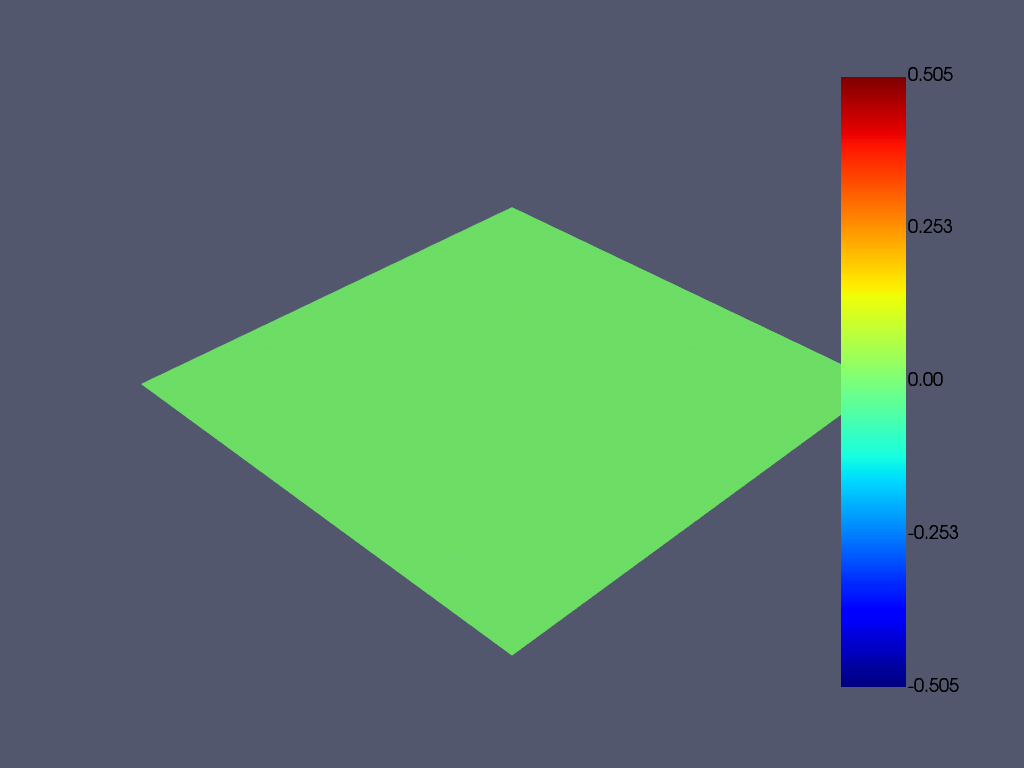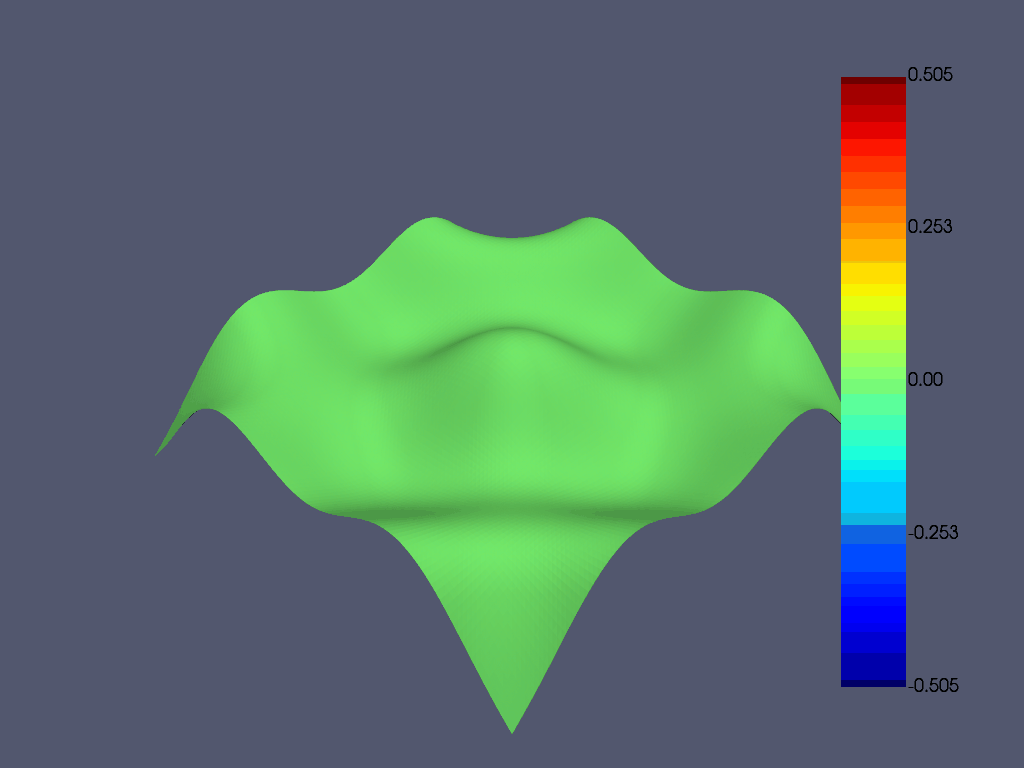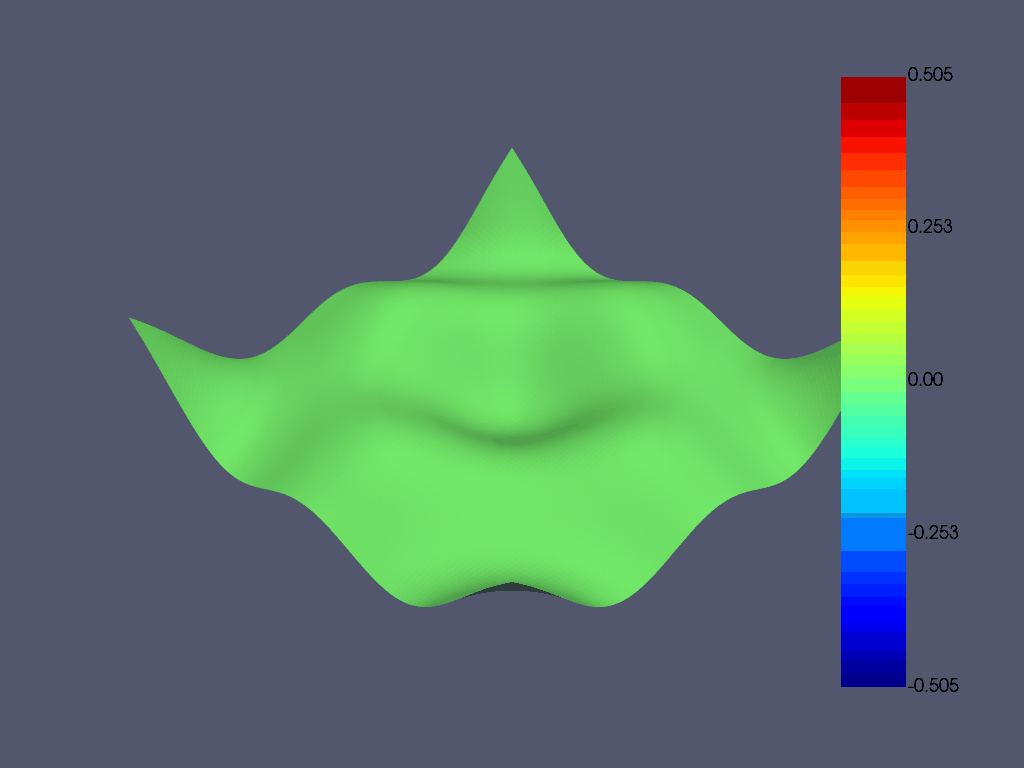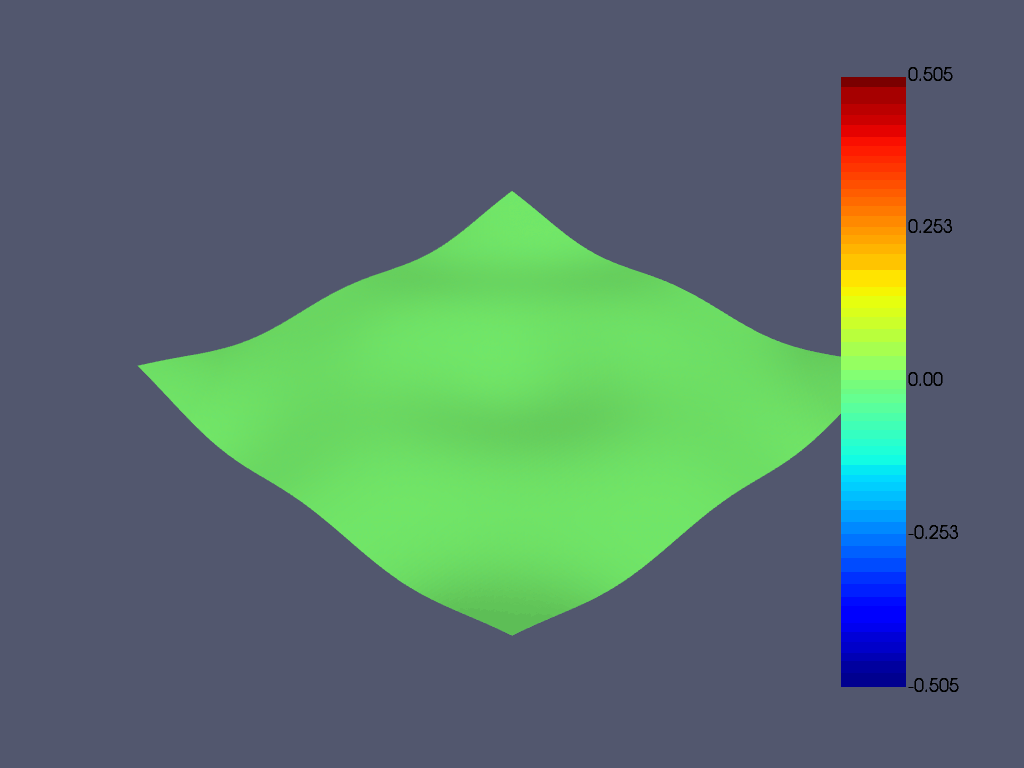
在 pyansys 中加载结果文件并绘制第 8 阶模态。
result = mapdl.result
print(result)
result.plot_nodal_displacement(7, show_displacement=True, displacement_factor=0.4)
PyMAPDL Result
Units : User Defined
Version : 23.1
Cyclic : False
Result Sets : 20
Nodes : 10201
Elements : 10000
Available Results:
NSL : Nodal displacements
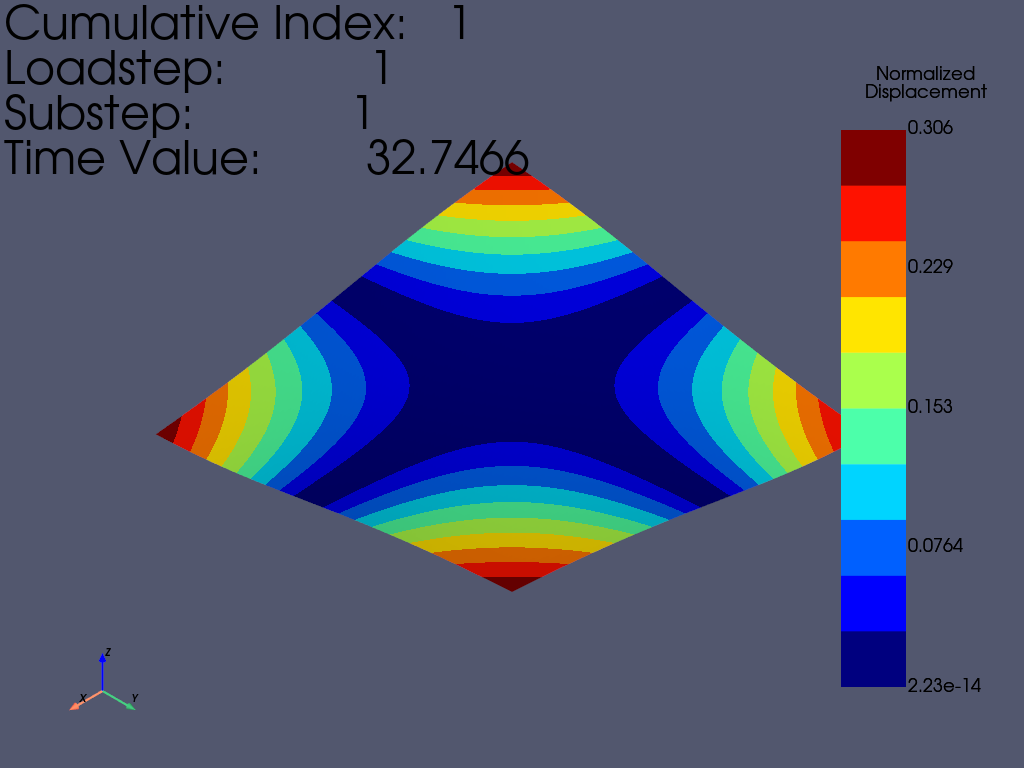
使用等高线绘制一阶模态
result.plot_nodal_displacement(
0, show_displacement=True, displacement_factor=0.4, n_colors=10
)
对模态分析结果进行动画处理
禁用 movie_filename ,并增加 n_frames 以获得更平滑的绘图。使用 loop=True 启用连续循环绘图。
result.animate_nodal_displacement(
18,
loop=False,
add_text=False,
n_frames=30,
displacement_factor=0.4,
show_axes=False,
background="w",
movie_filename="plane_vib.gif",
)
[(1.5773502691896262, 1.5773502691896262, 1.5773502691896262),
(0.0, 0.0, 0.0),
(0.0, 0.0, 1.0)]
Stop mapdl#
mapdl.exit()
Total running time of the script: (0 minutes 4.630 seconds)